Indo-Aryan
Regular Member
- Joined
- Jun 6, 2018
- Messages
- 815
- Likes
- 884
Congratulations boys!
Even Lord David Anthony is now proposing a Iranian homeland for PIE.
Even Lord David Anthony is now proposing a Iranian homeland for PIE.
https://archive.is/PULAk#selection-337.1554-337.1877Bluntness in speech, an unconcern about dress and appearance, a sense of equality within the fold that turned the village into " a collectivity of Patidar brothers" and a sense of superiority towards non-Patidars, a self-image of tough independent men... naturally given to ruling over others" marked the Patidar character.
Aren't modern Persians mainly J?Congratulations boys!
Even Lord David Anthony is now proposing a Iranian homeland for PIE.
Aren't modern Persians mainly J?



Modern Indians cluster away from the European cluster. Just like ancient Indians.
View attachment 128957
The Graveyard and the Buddhist shrine at Saidu Sharif I (Swat, Pakistan): fresh chronological and stratigraphic evidence https://www.researchgate.net/public...resh_chronological_and_stratigraphic_evidence
A Period of Acculturation in Ancient Gandhara - ResearchGate
Gandhara grave culture https://archive.is/3OyFB
View attachment 128958
Noteworthy Europeans with YDNA Haplogroup E1b1b - Page 16
Apricity is a European Cultural Communitywww.theapricity.com
| All 23 Roopkund_A samples and best qpAdm models. |
Sample ID | Gender | Y HG | p-Value | I8726 | Sintashta | Irula | WSHG | Mongolia_N | Tarim_EMBA |
I3351 | M | J2b2a~ | 0.8420 | 31% | 34% | 34% | | | |
I6938 | F | - | 0.2296 | 27% | 29% | 38% | | | 6% |
I2872 | F | - | 0.0772 | 41% | 28% | 31% | | | |
I3406 | M | J2a1a1b1 | 0.1210 | 47% | 26% | 15% | 4% | 10% | |
I6943 | M | J2b2a2b~ | 0.8163 | 37% | 24% | 39% | | | |
I2871 | F | - | 0.1623 | 44% | 23% | 33% | | | |
I3349 | F | - | 0.3871 | 31% | 22% | 47% | | | |
I3346 | M | E1b1b1b2a1a~ | 0.0375 | 32% | 22% | 47% | | | |
I6945 | F | - | 0.0743 | 10% | 20% | 70% | | | |
I6934 | F | - | 0.4091 | 54% | 18% | 28% | | | |
I6944 | F | - | 0.3330 | 21% | 18% | 61% | | | |
I3352 | M | R2a2b1b2b | 0.1488 | 48% | 15% | 37% | | | |
I7036 | M | R2 | 0.0670 | 18% | 14% | 59% | | 10% | |
I3344 | F | - | 0.0945 | 45% | 11% | 44% | | | |
I3343 | F | - | 0.2011 | 28% | 11% | 61% | | | |
I3402 | M | H3b | 0.0067 | 41% | 11% | 33% | | 15% | |
I6941 | M | R2 | 0.1235 | 8% | 10% | 74% | | 8% | |
I3342 | M | H1a1a4b | 0.1230 | 29% | 9% | 53% | | 10% | |
I7035 | F | - | 0.1260 | 20% | 6% | 74% | | | |
I2868 | M | H1a1b1 | 0.2126 | 20% | 6% | 74% | | | |
I3407 | M | H1a1a4b | 0.0880 | 15% | 4% | 81% | | | |
I6946 | M | R1a1a | 0.2905 | | | 100% | | | |
I6942 | M | R1a1a1b2a1a1a1f~ | 0.1593 | | | 100% | | | |
During the Expansion of Gujjars empire in 6th. and 7th. Century the Gujjars moved their capital from Ujjain to Rajor and Rajor to Kannauj.
It is written as highest caste in 9th. century by many Arab historians placed above than Brahmins and many valiant brave Gujjars were posted in Pamer region of Afghanistan to Check the invaders which they valiantly did for three centuries.
That is the reason that you will find lot Gujjars in present day Pakistan and Afghanistan sharing the same gotras as we have and speaking the same language Gojari.
In Afghanistan The Gujjars are written as Guzars and present Police Chief of Kabul is a Gujjar. It si reported by many newpapers that the fastest bowler of the world Shoaib Akhtar is a Kasana Gujjar. Hence the information provided by you about the Maharashtra Gujars is not read in DElhi and rajasthan only but in Pakistan and Afghanistan as well.
Lewa and Kharis said to be originated from Lava and Kushas are said to be the original gotras of Gujjars and many gotras are generated from these two, some of them named by the name of place they were rulers, others were named by some legendry warriors of this tribe.
Historically the term “Gurjara Pratihar” denoted a branch or group known as Bargujar Rajputs as identified by historians Prof Kielhorn and BD Chattopadhyay. They were feudatories of the Imperial Pratihars and very distinct from them.
Some information about "Leva Gurjars" in Maharasta | www.gurjarsonlin…
archived 3 Jan 2022 05:59:28 UTCarchive.is
View attachment 129236









The cause for its inaccuracy may be due to the fact that Mapmygenome has only collected DNA samples from 3,000 customers, most of whom are probably of Indian descent. The company may just not have collected enough samples from a diverse enough range of customers to be able to get certain people’s ancestry results right.
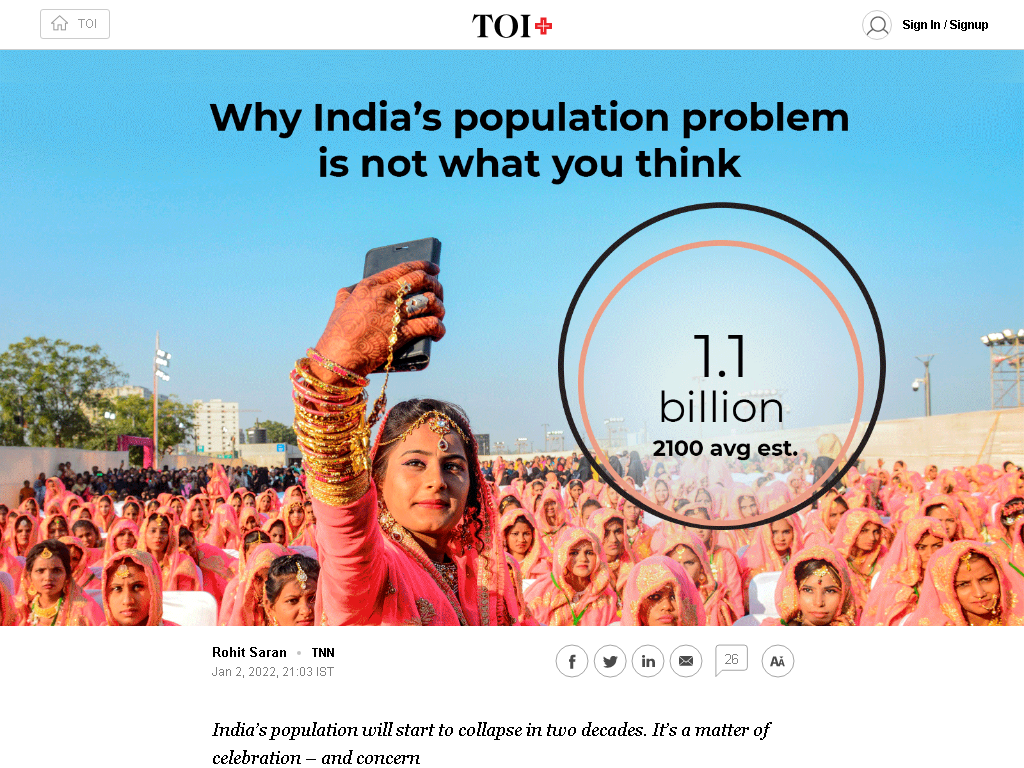
India may face a population implosion


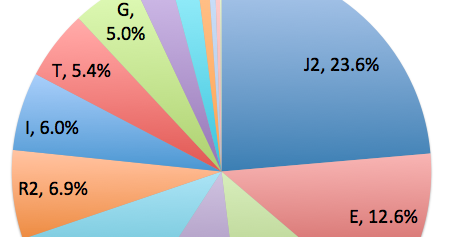
Haplogroup J is common in India, but E is very rare. American Journal of Physical Anthropology 145(1):21-9
R1a L657 in Roopkund lake, Uttarakhand from 800CE, but no Steppe
What does R-L657 in Roopkund lake remains mean?a-genetics.blogspot.com
All 23 Roopkund_A samples and best qpAdm models.
The E1b1b1a-M78 chromosome was observed in all the Roma groups studied and in the Hungarian reference population, but it was absent from Malaysian Indians and almost all Indian populations previously investigated.
| Thread starter | Similar threads | Forum | Replies | Date |
|---|---|---|---|---|
| N | lets talk about, Aryan invasion/Migrantion | History & Culture | 1 | |
|
|
Aryan Invasion Theory. Do you approve? | Subcontinent & Central Asia | 2 | |
|
|
Indo-Aryans vs Iranians | History & Culture | 5 | |
| P | European Misappropriation of Sanskrit led to the Aryan Race Theory | History & Culture | 2 |
